QuagmiR
Sequencing Analysis IsomiRs Identification
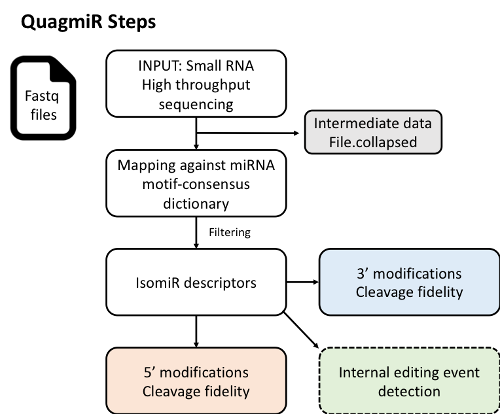
QuagmiR - the first cloud-based tool to analyze isomiRs from next generation sequencing data. Using a novel and flexible searching algorithm designated for the detection and annotation of isomiRs of heterogeneous nature. It permits extensive customization of the query process and reference databases to meet the user’s diverse demands. QuagmiR is written in Python and can be obtained freely from Github. QuagmiR can be run from command-line on local machines, as well as high performance servers. A web-accessible version of the tool has also been made available for use by academic researchers through the National Cancer Institute-funded Seven Bridges Cancer Genomics Cloud.
Homepage: Link
Reference:
[Journal]
Organism Specific:
Reference Genome Needed:
Online/Local:
/
Installation/User Level: easy
User Adjustability:
User support:
Precomputed Target Results Available For Download:
Input Data Required: